Sequence Database Setup: MSIPI
| SwissProt Release 56.0 |
| MSIPI includes a number of SwissProt entries representing common contaminants.
As a result, it was affected by the change in the syntax of the Fasta title line in SwissProt
release 56.0 (UniProtKB 14.0). This page has been updated to illustrate the new parse rules.
|
|
Overview
MSIPI
is a database derived from IPI that contains additional information
about cSNPs, N-terminus peptides, and known variants in a format suitable for mass spectrometry
search engines. MSIPI is produced by the
Max-Planck Institute for
Biochemistry at Martinsried
and the University of Southern Denmark,
and distributed by the EBI.
There are currently two MSIPI databases, Human and Mouse. This document uses the Human database as an example.
To work with the Mouse database, simply substitute the word "MOUSE" for "HUMAN".
Download
ftp://ftp.ebi.ac.uk/pub/databases/IPI/msipi/current/
for the latest release.
ftp://ftp.ebi.ac.uk/pub/databases/IPI/msipi/old/
for earlier releases.
There are two files: a Fasta database file, (msipi.HUMAN_slim.fasta.gz), and file that also includes
the reversed sequences, (msipi.HUMAN_decoy_slim.fasta.gz), providing a ready-made, concatenated
decoy database. The inclusion of the word slim in the file name
indicates that the title has been abbreviated.
There isn't an easy way to configure a local full text reference file because MSIPI has a number
of Swiss-Prot sequences appended to the end, representing common contaminants. Mascot requires a 1:1
correspondence between the Fasta file and the full text reference file, so the standard IPI *.dat file
cannot be used. Fortunately, you can query an SRS server to get full text reports, as shown in the
configuration example below.
Use of code letter J
As described in the Nature
Methods paper, the additional peptide sequences in MSIPI, representing cSNPs, N-terminus peptides, and
known variants, are delimited from the original protein sequences using the letter J. This code letter
is not used in a default Mascot configuration, and has a mass of zero. You need to add this letter to any
enzyme definitions that you plan to use with MSIPI as an additional cleavage site. To avoid confusion,
create new definitions with self-explanatory names, such as TrypsinMSIPI.
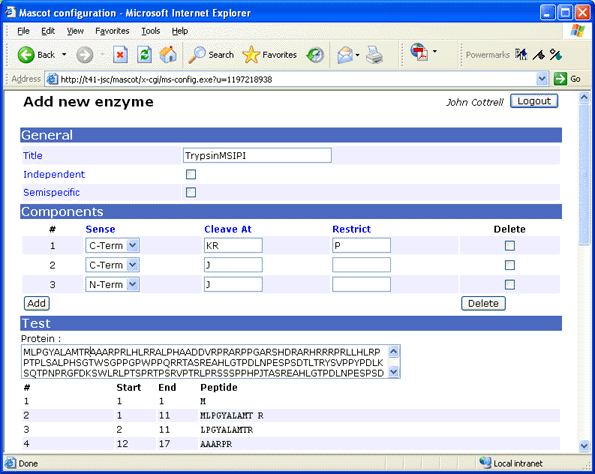
For earlier versions of Mascot, use a text editor to modify the enzymes configuration
file:
Title:TrypsinMSIPI
Cleavage[0]:KR
Restrict[0]:P
Cterm[0]
Cleavage[1]:J
Cterm[1]
Cleavage[2]:J
Nterm[2]
*
Title:TrypsinMSIPI/P
Cleavage[0]:KRJ
Cterm[0]
Cleavage[1]:J
Nterm[1]
*
The supplementary
material to the Nature Methods paper suggests setting the mass of J to that of Asparagine, (114).
However, the letter J is sometimes used as the ambiguity code for
I
or L. If you search databases in which J is used in this way, you will need to set the
mass of J to 113. This will mean that, when searching MSIPI with missed cleavages set
greater than zero, you will sometimes see spurious peptide sequences containing J among the low scoring,
random matches.
Otherwise,
best to leave the mass of J at zero. You will still see peptide sequences containing J
in the result reports, but this will not change any of the mass values or the matches.
Taxonomy
Taxonomy is not required because all entries are from the same species
Parse Rules
Typical Fasta title lines from the non-decoy Fasta files are:
>MSIPI:IPI00000001.2|
Gene_Symbol=STAU1 Isoform Long of Double-stranded
RNA-binding protein Staufen homolog 1 lng=577 # CON[595,R,359,A] #
>MSIPI:sp|P07758|A1AT1_MOUSE
Alpha-1-antitrypsin 1-1 OS=Mus musculus GN=Serpina1a PE=1 SV=4
The parse rule must be general enough to match both the IPI entries and the Swiss-Prot
contaminant entries.
Accession from Fasta title: ">MSIPI:s*p*|*\([^| .]*\)"
Description from Fasta title: ">[^ ]* \(.*\)"
These rules will not work for the Fasta file that includes concatenated decoy entries. It doesn't appear possible for a single
parse rule to extract accession strings for the target entries that will be recognised by an SRS server and
still extract unique accessions for the reversed SwissProt sequences.
>MSIPI:IPI00000001.2| Gene_Symbol=STAU1 ...
>MSIPI:REV00000001.2| Gene_Symbol=STAU1 ...
>MSIPI:sp|P07758|A1AT1_MOUSE Alpha-1-antitrypsin ...
>MSIPI:REVsp|P07758|A1AT1_MOUSE Alpha-1-antitrypsin ...
The following rule will extract a unique accession string, but the link for Protein View will fail:
Accession from Fasta title: ">MSIPI:\([^ .]*\)"
Configuration
For this example, the Fasta file was downloaded to
C:\sequence\MSIPI_human\current,
decompressed using gzip,
and renamed to MSIPI_human_3.48.fasta.
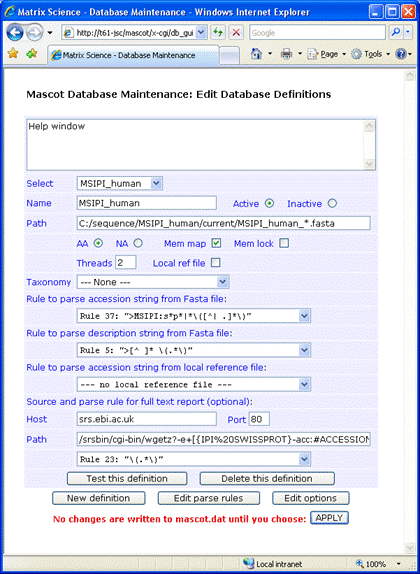
As mentioned earlier, there isn't a local reference file corresponding to the Fasta. Full text for individual
entries can be retrieved from an SRS server, such as the public server at EBI. The following syntax for the
Path field retrieves entries from both IPI and Swiss-Prot:
/srsbin/cgi-bin/wgetz?-e+[{IPI%20SWISSPROT}-acc:#ACCESSION#]+-vn+2
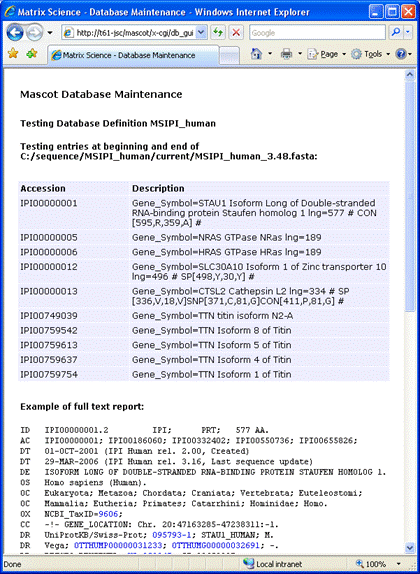
If you get the message Error: Nothing returned by call to ... instead of the sample
full text report, this usually indicates that
the Mascot Server cannot access the Internet because of a firewall or proxy server problem. In the case of a proxy server,
the host, port and, if necessary, a user name and password are defined in mascot.dat using the labels proxy_server, proxy_username,
and proxy_password. Choose Edit Options in Database Maintenance. For example:
proxy_server http://my_proxy:3128
If you don't require full text in a Mascot Protein View report, simply leave the Host, Port, and Path fields blank
and choose
--- no full text report ---
in the drop down list.
Always test a new definition before applying the changes to mascot.dat.
|